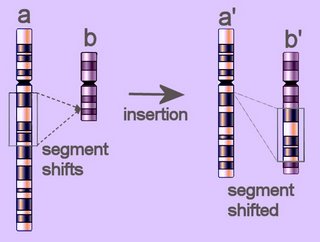
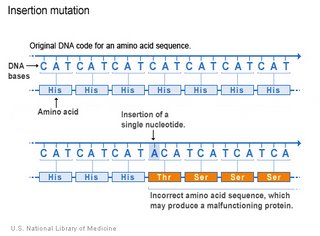
Duplication of a segment of the arm of a chromosome is similar to
insertion of sequences at the level of genes. Duplications have played an important role in the
evolution of the
genomes of many species.
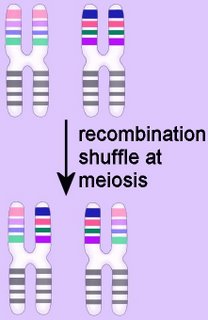
Duplication results from unequal
recombination (crossing-over) when homologous chromosomes are mis-aligned during
meiosis. The frequency of duplication events depends upon the number of shared repetitive elements on the homologous chromosomes. The recombination products of duplication events include duplication at the site of exchange combined with a reciprocal deletion. Thus, duplication events produce an increase of some genetic material. Once established in a genome,
functional gene duplicates persist by a variety of mechanisms.
In 1970, Susumu
Ohno proposed that several rounds of genome duplication (polyploidy) occurred during vertebrate evolution (see
Skrabanek and Wolfe, 1998). Ohno hypothesized that “natural selection merely modified, while redundancy created”, that is, while mutation merely provides material for possible selection, duplication creates. Ohno further hypothesized that evolutionary leaps such as the emergence of new body plans would require major events, as drastic as whole-genome duplication.[
r] Study of evolutionary divergence rates in duplicated genes, and their comparison with their non-duplicated counterparts have demonstrated that "Ohno's model is correct in 95% of cases of accelerated divergence: one gene copy preserved the ancestral function, while the other copy was free to diverge. The new functions included gene silencing, anti-viral defense mechanisms, and new signaling pathways. Moreover, as a group, the ancestral and derived function genes show distinct patterns in expression, localization, and generality of function."[
s]
Comparison of the tetraodon and human genomes has revealed a signature of duplicate mapping. Studying the gene correspondence between human genes and their orthologs in fish chromosomes, a team of researchers demonstrated that the human genome was doubly covered by interleaved stretches of homology to pairs of fish chromosomes, revealing that the signature of whole-genome duplication exists in the fish genome, pointing to an event common to zebrafish, fugu, and pufferfish. These results illustrate that the signatures of evolutionary change were in fact applicable across kingdoms, exhibiting basic principles of evolutionary mechanisms.[
s] Comparative maps of mammalian genomes show that large chunks of chromosome are
conserved between species. For example, about 200 conserved chromosomal regions (synteny) are known between mouse and human.[
s]
The flowering plants, angiosperms have a marked tendency to undergo chromosomal duplication ('polyploidization') and subsequent gene loss ('diploidization').[
s]
Genome sequencing suggests that the genome of yeast duplicated (became polyploid) at some point during its evolutionary past. Genome duplication in an ancestor of
Saccharomyces cerevisiae occurred relatively recently, following its divergence from many other fungi such as
Kluyveromyces lactis. It has been suggested that duplicated chromosomal regions in yeast are remnants of a whole-genome duplication (tetraploidy) that occurred 10^8 years ago [
s]. Subsequently, many of the duplicated genes were deleted and the chromosomes were rearranged by reciprical translocation. See
Yeast Gene Duplications website for details of the duplicated chromosomal regions.
Modeling gene and genome duplications in eukaryotes. modified:
Recent analysis of complete eukaryotic genome sequences has revealed that gene duplication has been rampant.
Moreover, next to a continuous mode of gene duplication, in many eukaryotic organisms the complete genome has been duplicated in their evolutionary past.
Such large-scale gene duplication events have been associated with important evolutionary transitions or major leaps in development and adaptive radiations of species. Here, we present an evolutionary model that simulates the duplication dynamics of genes, considering genome-wide duplication events and a continuous mode of gene duplication. Modeling the evolution of the different functional categories of genes assesses the importance of different duplication events for gene families involved in specific functions or processes. By applying our model to the Arabidopsis genome, for which there is compelling evidence for three whole-genome duplications, we show that gene loss is strikingly different for large-scale and small-scale duplication events and highly biased toward certain functional classes. We provide evidence that some categories of genes were almost exclusively expanded through large-scale gene duplication events. In particular, we show that the three whole-genome duplications in Arabidopsis have been directly responsible for >90% of the increase in
transcription factors,
signal transducers, and
developmental genes in the last 350 million years. Our evolutionary model is widely applicable and can be used to evaluate different assumptions regarding small- or large-scale gene duplication events in eukaryotic genomes.
Maere S,
De Bodt S,
Raes J,
Casneuf T,
Van Montagu M,
Kuiper M,
Van de Peer Y.
Modeling gene and genome duplications in eukaryotes. (Free Full Text Article)
Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5454-9. Epub 2005 Mar 30.
Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity.
Controversy surrounds the apparent rising maximums of morphological complexity during eukaryotic evolution, with organisms increasing the number and nestedness of developmental areas as evidenced by morphological elaborations reflecting area boundaries. No "predictable drive" to increase this sort of complexity has been reported. Recent genetic data and theory in the general area of gene dosage effects has engendered a robust "gene balance hypothesis," with a theoretical base that makes specific predictions as to gene content changes following different types of gene duplication. Genomic data from both chordate and angiosperm genomes fit these predictions: Each type of duplication provides a one-way injection of a biased set of genes into the gene pool. Tetraploidies and balanced segments inject bias for those genes whose products are the subunits of the most complex biological machines or cascades, like
transcription factors (TFs) and
proteasome core proteins. Most duplicate genes are removed after tetraploidy. Genic balance is maintained by not removing those genes that are dose-sensitive, which tends to leave duplicate "functional modules" as the indirect products (spandrels) of purifying selection. Functional modules are the likely precursors of coadapted gene complexes, a unit of natural selection. The result is a predictable drive mechanism where "drive" is used rigorously, as in "meiotic drive." Rising morphological gain is expected given a supply of duplicate functional modules. All flowering plants have survived at least three large-scale duplications/diploidizations over the last 300 million years (Myr). An equivalent period of tetraploidy and body plan evolution may have ended for animals 500 million years ago (Mya). We argue that "balanced gene drive" is a sufficient explanation for the trend that the maximums of morphological complexity have gone up, and not down, in both plant and animal eukaryotic lineages.
Freeling M,
Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity.
Genome Res. 2006 Jul;16(7):805-14.
Modeling gene and genome duplications in eukaryotes. [Proc Natl Acad Sci U S A. 2005] PMID: 15800040
Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. [Genome Res. 2006] PMID: 16760422
Widespread genome duplications throughout the history of flowering plants. [Genome Res. 2006] PMID: 16702410
New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. [Genome Res. 2003] PMID: 12799346
Extensive genomic duplication during early chordate evolution. [Nat Genet. 2002] PMID: 12032567
See all Related Articles...Extensive genomic duplication during early chordate evolution.
Opinions on the hypothesis that ancient genome duplications contributed to the vertebrate genome range from strong skepticism to strong credence. Previous studies concentrated on small numbers of gene families or chromosomal regions that might not have been representative of the whole genome, or used subjective methods to identify paralogous genes and regions. Here we report a systematic and objective analysis of the draft human genome sequence to identify paralogous chromosomal regions (paralogons) formed during chordate evolution and to estimate the ages of duplicate genes. We found that the human genome contains many more paralogons than would be expected by chance. Molecular clock analysis of all protein families in humans that have orthologs in the fly and nematode indicated that a burst of gene duplication activity took place in the period 350 650 Myr ago and that many of the duplicate genes formed at this time are located within paralogons. Our results support the contention that many of the gene families in vertebrates were formed or expanded by large-scale DNA duplications in an early chordate. Considering the incompleteness of the sequence data and the antiquity of the event, the results are compatible with at least one round of polyploidy.
McLysaght A,
Hokamp K,
Wolfe KH. Extensive genomic duplication during early chordate evolution.
Nat Genet. 2002 Jun;31(2):200-4. Epub 2002 May 28. Comment in:
Nat Genet. 2002 Jun;31(2):128-9.Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. [Mol Biol Evol. 2004] PMID: 15014147
New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. [Genome Res. 2003] PMID: 12799346
Ancient large-scale genome duplications: phylogenetic and linkage analyses shed light on chordate genome evolution. [Mol Biol Evol. 1998] PMID: 9729879
Phylogenetic analyses alone are insufficient to determine whether genome duplication(s) occurred during early vertebrate evolution. [J Exp Zoolog B Mol Dev Evol. 2003] PMID: 14508816
Phylogenetic analysis of T-Box genes demonstrates the importance of amphioxus for understanding evolution of the vertebrate genome. [Genetics. 2000] PMID: 11063699
See all Related Articles...Timing and mechanism of ancient vertebrate genome duplications -- the adventure of a hypothesis.
Complete genome doubling has long-term consequences for the genome structure and the subsequent evolution of an organism. It has been suggested that two genome duplications occurred at the origin of vertebrates (known as the 2R hypothesis). However, there has been considerable debate as to whether these were two successive duplications, or whether a single duplication occurred, followed by large-scale segmental duplications. In this article, we review and compare the evidence for the 2R duplications from vertebrate genomes with similar data from other more recent polyploids.
Panopoulou G,
Poustka AJ. Timing and mechanism of ancient vertebrate genome duplications -- the adventure of a hypothesis.
Trends Genet. 2005 Oct;21(10):559-67
Evolution and diversity of fish genomes. [Curr Opin Genet Dev. 2003] PMID: 14638319
Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. [Proc Natl Acad Sci U S A. 2004] PMID: 14757817
Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. [Mol Biol Evol. 2004] PMID: 15014147
Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. [Nature. 2004] PMID: 15496914
See all Related Articles...Turning the clock back on ancient genome duplication. [Curr Opin Genet Dev. 2003] PMID: 14638327
Were vertebrates octoploid? [Philos Trans R Soc Lond B Biol Sci. 2002] PMID: 12028790
Phylogenetic dating and characterization of gene duplications in vertebrates: the cartilaginous fish reference. [Mol Biol Evol. 2004] PMID: 14694077
Phylogenetic analyses alone are insufficient to determine whether genome duplication(s) occurred during early vertebrate evolution. [J Exp Zoolog B Mol Dev Evol. 2003] PMID: 14508816
Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. [Mol Biol Evol. 2002] PMID: 12200472
See all Related Articles...Tables
Mechanisms of Biological Evolution :
Gene Regulation in E.coli :
|













































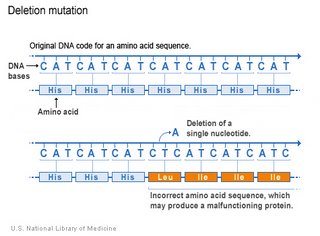
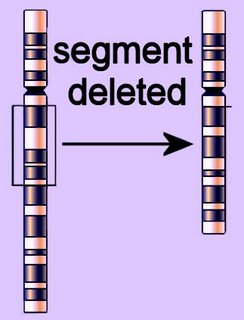 The deletion mutation eliminates one or more nucleotides from a DNA sequence, and may produce a non-functional protein. If the number of deleted bases is not a multiple of 3, then the deletion will cause frameshift, with potentially serious consequences.
The deletion mutation eliminates one or more nucleotides from a DNA sequence, and may produce a non-functional protein. If the number of deleted bases is not a multiple of 3, then the deletion will cause frameshift, with potentially serious consequences.